File:Fig2 Asiimwe JofTransMed21 19.png
Original file (1,961 × 1,653 pixels, file size: 703 KB, MIME type: image/png)
Summary
| Description |
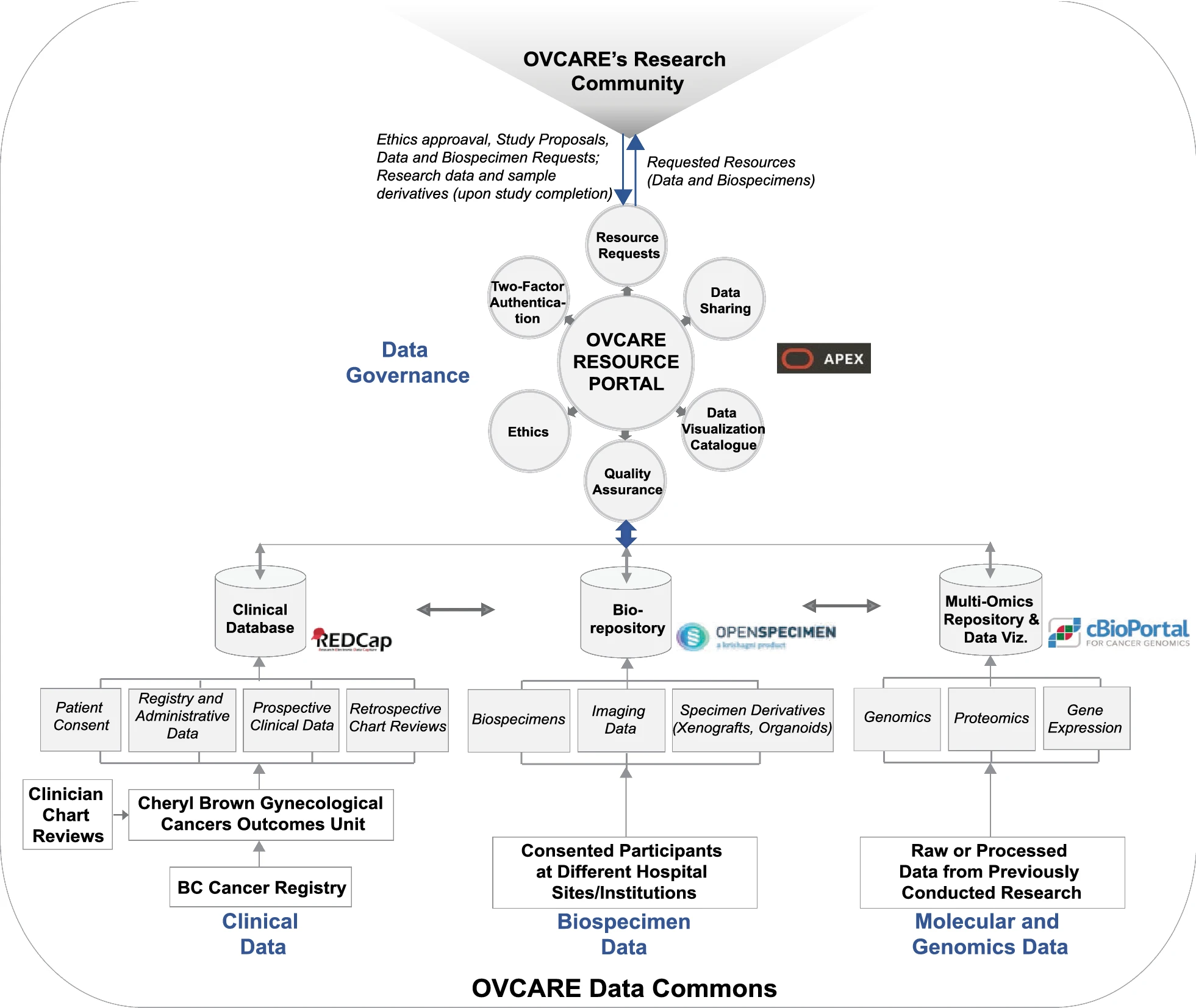
Figure 2. OVCARE’s data commons infrastructure and software stack. The overall data commons infrastructure comprises of five main components: (1) A clinical database (REDCap) that consolidates and manages clinical data collections from the BC Cancer Registry and the Cheryl Brown Gynecological Cancers Outcomes Unit; (2) a Library Information Management System (OpenSpecimen) that stores and manages biospecimens collected from consented participants at different hospital sites (i.e., Vancouver General Hospital, the University of British Columbia Hospital, BC Cancer Vancouver, and now a few more centers in BC); (3) the cBioPortal that supports the exploration, analysis, and visualization of clinical attributes and molecular profiles from patient tumor samples; (4) the OVCARE Resource Portal (ORP) that governs data and resource sharing based on stipulated protocols, SOPs, and research ethics; and (5) the Research Community (this includes the OVCARE internal research and informatics team, and the broader research community that OVCARE serves). Each of the components (REDCap, OpenSpecimen, cBioPortal, ORP) identified to meet our research needs are separately hosted in our hospital’s computing environment and programmatically interlinked through API calls. The data from the different domains are interlinked using system-wide unique identifiers that link patients to their biospecimen collections and molecular/genomics data. To access the amassed clinical and biospecimen collections, authenticated researchers in the OVCARE research community send data and sample acquisition requests to the ORP through which those requests are met by informatics staff, if all stipulated requirements including ethics approval are met. Upon successful data and sample acquisition, researchers conduct their respective studies, and the data generated (raw or processed, and/ biospecimen derivatives) from their research are retuned to OVCARE, making it available for re-purposing/secondary use. Furthermore, molecular data returned to the data commons are linked back to the available and stored patient biospecimens. Together with clinical outcomes, these molecular profiles are further explored, analyzed, and visualized using the cBio Cancer Genomics Portal. |
|---|---|
| Source |
Asiimwe, R.; Lam, S.; Leung, S.; Wang, S.; Wan, R.; Tinker, A.; McAlpine, J.N.; Woo, M.M.M.; Huntsman, D.G.; Talhouk, A. (2021). "From biobank and data silos into a data commons: Convergence to support translational medicine". Journal of Translational Medicine 19: 493. doi:10.1186/s12967-021-03147-z. |
| Date |
2021 |
| Author |
Asiimwe, R.; Lam, S.; Leung, S.; Wang, S.; Wan, R.; Tinker, A.; McAlpine, J.N.; Woo, M.M.M.; Huntsman, D.G.; Talhouk, A. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 4.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 16:48, 5 January 2022 |  | 1,961 × 1,653 (703 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following 2 pages use this file:







