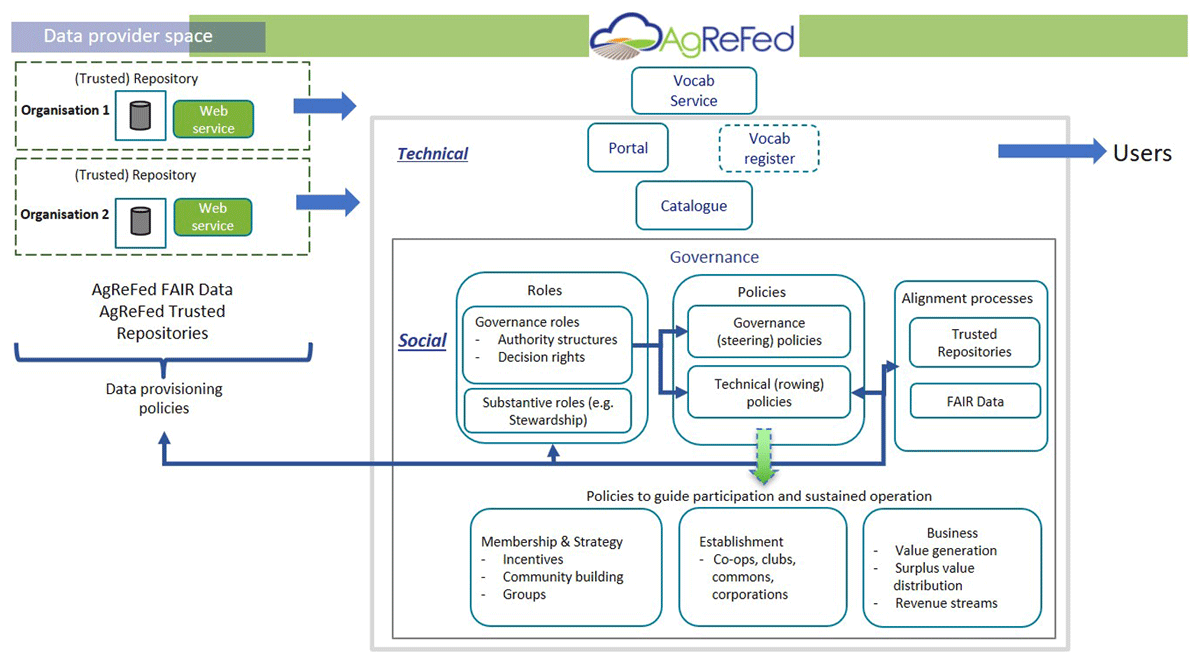
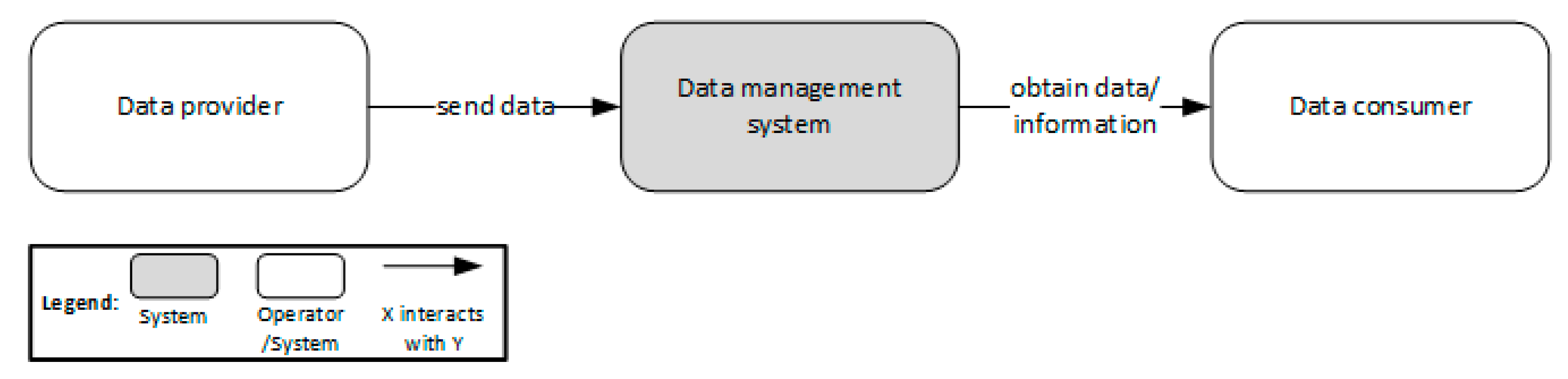
Difference between revisions of "Main Page/Featured article of the week/2023"
Shawndouglas (talk | contribs) (Added last week's article of the week) |
Shawndouglas (talk | contribs) (Added last week's article of the week) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 20–27:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 03–09:</h2> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Foster JoMedLibAss22 110-2.png|240px]]</div> | |||
'''"[[Journal:Implementing an institution-wide electronic laboratory notebook initiative|Implementing an institution-wide electronic laboratory notebook initiative]]"''' | |||
To strengthen institutional research [[Information management|data management]] practices, the [[Indiana University – Purdue University Indianapolis|Indiana University School of Medicine]] (IUSM) licensed an [[electronic laboratory notebook]] (ELN) to improve the organization, security, and shareability of [[information]] and data generated by the school’s researchers. The Ruth Lilly Medical Library led implementation on behalf of the IUSM’s Office of Research Affairs. This article describes the pilot and full-scale implementation of an ELN at IUSM. The initial pilot of the ELN in late 2018 involved 15 research labs, with access expanded in 2019 to all academic medical school constituents ... ('''[[Journal:Implementing an institution-wide electronic laboratory notebook initiative|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 27–April 02:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Ronalter EnviroDevSust22 660.png|240px]]</div> | |||
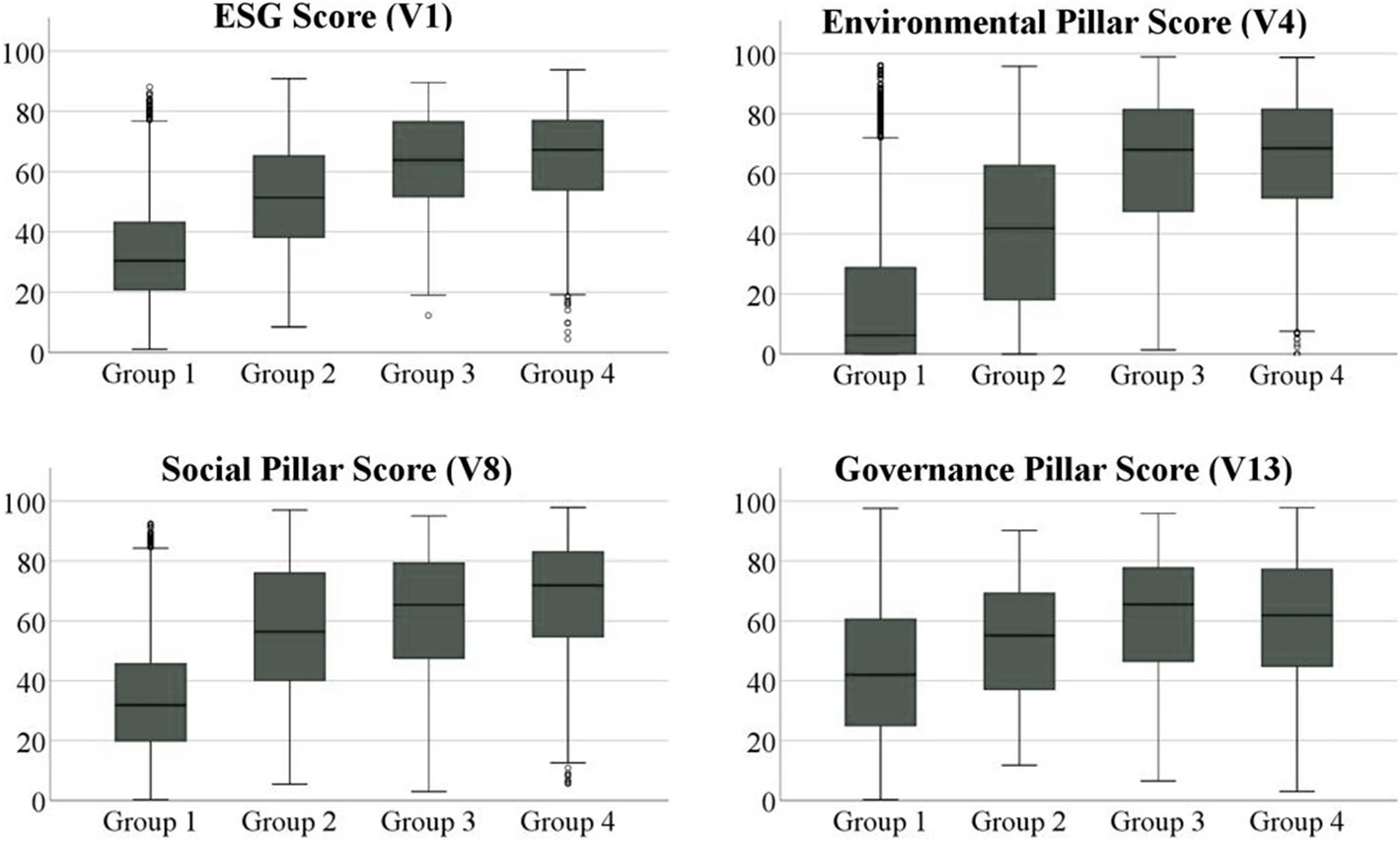
'''"[[Journal:Quality and environmental management systems as business tools to enhance ESG performance: A cross-regional empirical study|Quality and environmental management systems as business tools to enhance ESG performance: A cross-regional empirical study]]"''' | |||
The growing societal and political focus on sustainability at the global level is pressuring companies to enhance their [[wikipedia:Environmental, social, and corporate governance|environmental, social, and governance]] (ESG) performance to satisfy respective stakeholder needs and ensure sustained business success. With a data sample of 4,292 companies from Europe, East Asia, and North America, this work aims to prove through a cross-regional empirical study that [[quality management system]]s (QMSs) and [[environmental management system]]s (EMSs) represent powerful business tools to achieve this enhanced ESG performance. Descriptive and cluster analyses reveal that firms with QMSs and/or EMSs accomplish statistically significant higher ESG scores than companies without such management systems. Furthermore, the results indicate that operating both types of management systems simultaneously increases performance in the environmental and social pillar even further, while the governance dimension appears to be affected mainly by the adoption of EMSs alone ... ('''[[Journal:Quality and environmental management systems as business tools to enhance ESG performance: A cross-regional empirical study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 20–26:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:GA Terlouw JofCheminfo22 14.png|240px]]</div> | |||
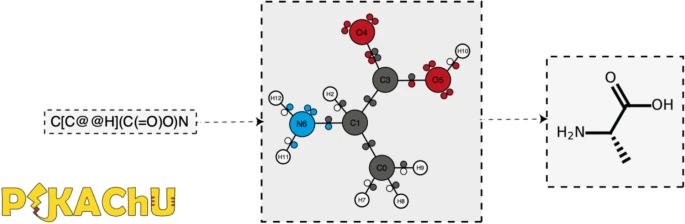
'''"[[Journal:PIKAChU: A Python-based informatics kit for analyzing chemical units|PIKAChU: A Python-based informatics kit for analyzing chemical units]]"''' | |||
As efforts to computationally describe and simulate the biochemical world become more commonplace, computer programs that are capable of ''in silico'' chemistry play an increasingly important role in biochemical research. While such programs exist, they are often dependency-heavy, difficult to navigate, or not written in [[Python (programming language)|Python]], the programming language of choice for [[Bioinformatics|bioinformaticians]]. Here, we introduce PIKAChU (Python-based Informatics Kit for Analysing CHemical Units), a [[Chemical informatics|cheminformatics]] toolbox with few dependencies implemented in Python. PIKAChU builds comprehensive molecular graphs from simplified molecular-input line-entry system (SMILES) strings, which allow for easy downstream analysis and visualization of molecules. ... ('''[[Journal:PIKAChU: A Python-based informatics kit for analyzing chemical units|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 13–19:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:GA Karim JofKSUScience2022 34-2.jpg|240px]]</div> | |||
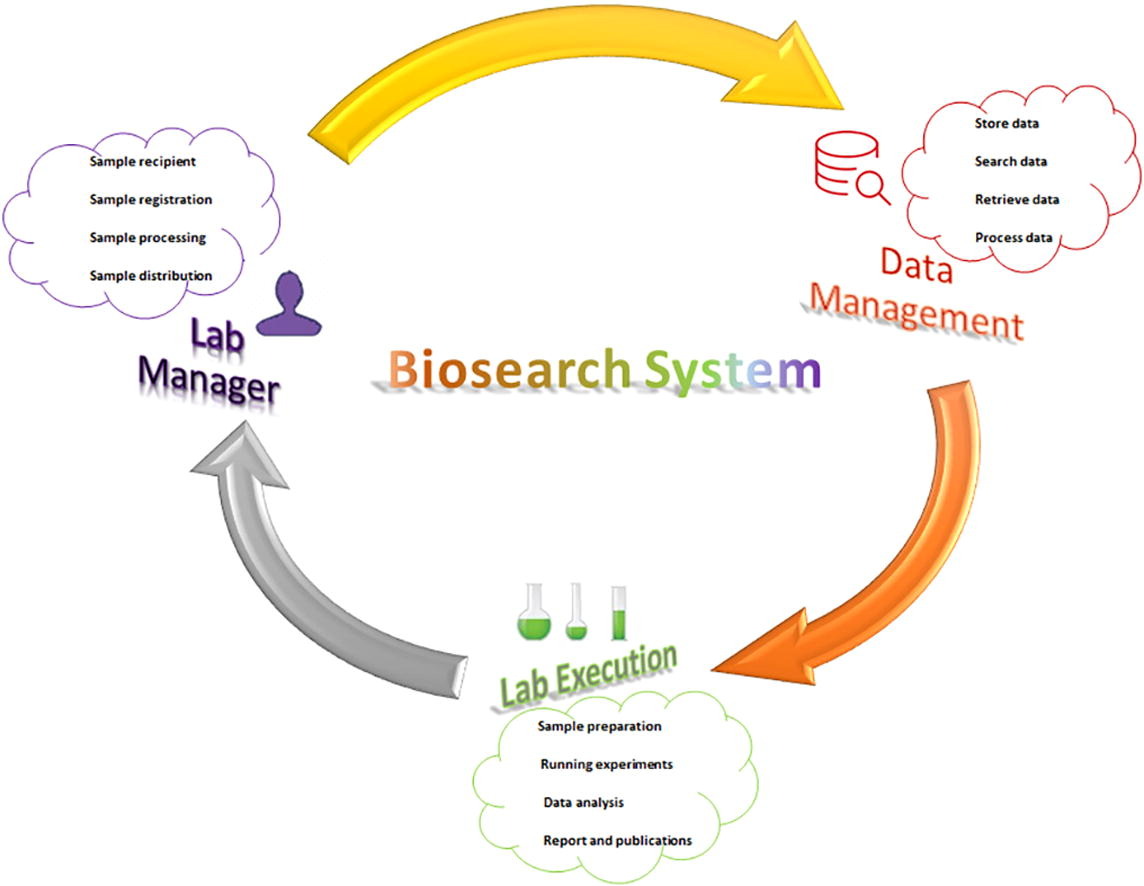
'''"[[Journal:Development of Biosearch System for biobank management and storage of disease-associated genetic information|Development of Biosearch System for biobank management and storage of disease-associated genetic information]]"''' | |||
Databases and software are important to manage modern high-throughput [[Laboratory|laboratories]] and store clinical and [[Genome informatics|genomic information]] for [[quality assurance]]. Commercial software is expensive, with proprietary code issues, while academic versions have adaptation issues. Our aim was to develop an adaptable in-house software system that can store specimen- and disease-associated genetic information in [[biobank]]s to facilitate [[translational research]]. A prototype was designed per the research requirements, and computational tools were used to develop the software under three tiers, using Visual Basic and ASP.net for the presentation tier, SQL Server for the data tier, and Ajax and JavaScript for the business tier. We retrieved specimens from the biobank using this software and performed microarray-based transcriptomic analysis to detect differentially expressed genes (DEGs) ... ('''[[Journal:Development of Biosearch System for biobank management and storage of disease-associated genetic information|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 06–12:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Andrés-Hernández FrontNut2022 9.jpg|240px]]</div> | |||
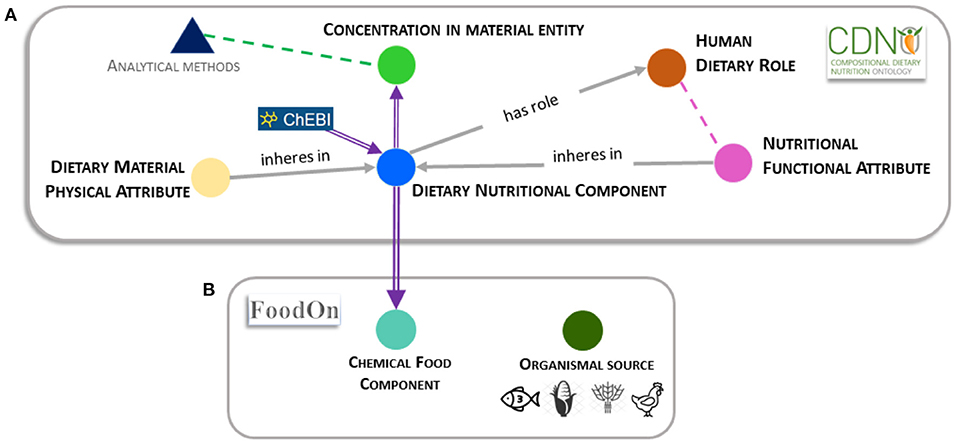
'''"[[Journal:Establishing a common nutritional vocabulary: From food production to diet|Establishing a common nutritional vocabulary: From food production to diet]]"''' | |||
Informed policy and decision-making for food systems, [[Food security|nutritional security]], and global health would benefit from standardization and comparison of food composition data, spanning production to consumption. To address this challenge, we present a formal [[controlled vocabulary]] of terms, definitions, and relationships within the [https://www.cdno.info/ Compositional Dietary Nutrition Ontology] (CDNO) that enables description of nutritional attributes for material entities contributing to the human diet. We demonstrate how ongoing community development of CDNO classes can harmonize trans-disciplinary approaches for describing [[Nutritional science|nutritional components]] from food production to diet ... ('''[[Journal:Establishing a common nutritional vocabulary: From food production to diet|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 28–March 05:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Melanitis MATECWebConf21 349.png|240px]]</div> | |||
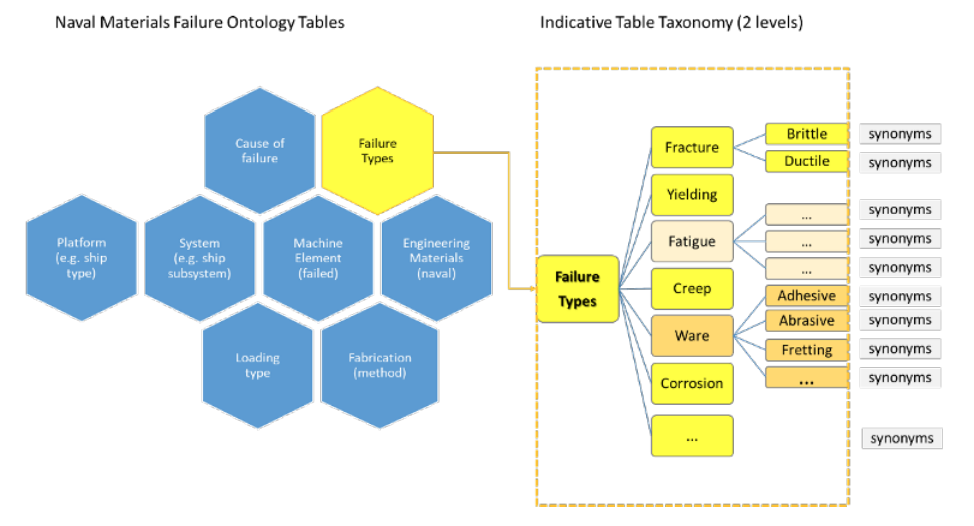
'''"[[Journal:Designing a knowledge management system for naval materials failures|Designing a knowledge management system for naval materials failures]]"''' | |||
Implemented materials fail from time to time, requiring [[failure analysis]]. This type of scientific analysis expands into [[Forensic science|forensic]] engineering for it aims not only to identify individual and symptomatic reasons for failure, but also to assess and understand repetitive failure patterns, which could be related to underlying material faults, design mistakes, or maintenance omissions. Significant [[information]] can be gained and studied from carefully documenting and managing the data that comes from failure analysis of materials, including in the naval industry. The NAVMAT research project, presented herein, attempts an interdisciplinary approach to [[materials informatics]] by integrating materials engineering and [[Informatics (academic field)|informatics]] under a platform of [[Information management|knowledge management]]. Our approach utilizes a focused, common-cause failure analysis methodology for the naval and marine environment. The platform's design is dedicated to the effective recording, efficient indexing, and easy and accurate retrieval of relevant information, including the associated history of maintenance and secure operation concerning failure incidents of marine materials, components, and systems in an organizational fleet. ... ('''[[Journal:Designing a knowledge management system for naval materials failures|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 20–27:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Yoon LabAniRes22 38.png|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Yoon LabAniRes22 38.png|240px]]</div> | ||
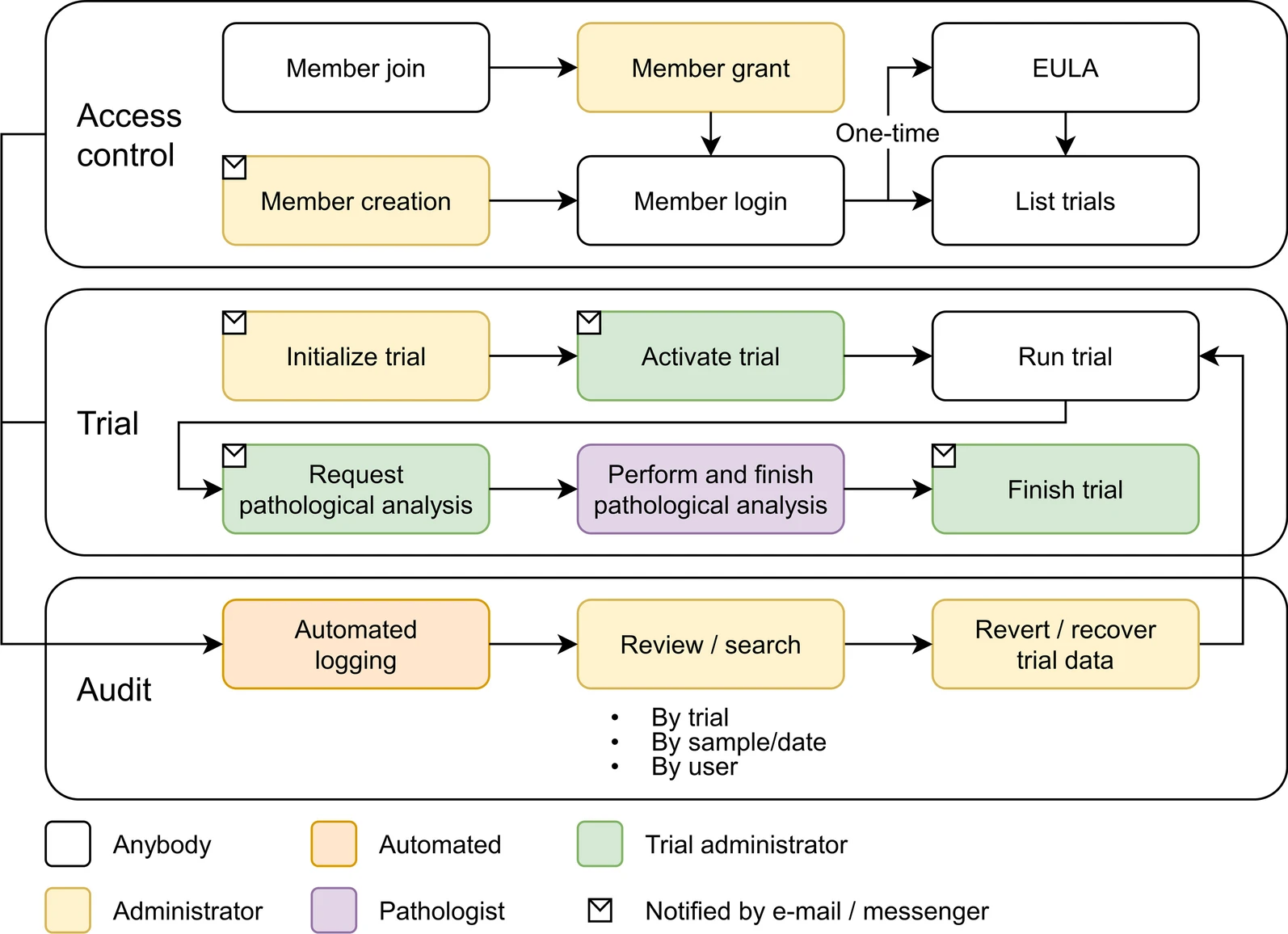
'''"[[Journal:Laboratory information management system for COVID-19 non-clinical efficacy trial data|Laboratory information management system for COVID-19 non-clinical efficacy trial data]]"''' | '''"[[Journal:Laboratory information management system for COVID-19 non-clinical efficacy trial data|Laboratory information management system for COVID-19 non-clinical efficacy trial data]]"''' | ||
Latest revision as of 17:02, 10 April 2023
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 - 2023 |
Featured article of the week archive - 2023
Welcome to the LIMSwiki 2023 archive for the Featured Article of the Week.
Featured article of the week: April 03–09:"Implementing an institution-wide electronic laboratory notebook initiative" To strengthen institutional research data management practices, the Indiana University School of Medicine (IUSM) licensed an electronic laboratory notebook (ELN) to improve the organization, security, and shareability of information and data generated by the school’s researchers. The Ruth Lilly Medical Library led implementation on behalf of the IUSM’s Office of Research Affairs. This article describes the pilot and full-scale implementation of an ELN at IUSM. The initial pilot of the ELN in late 2018 involved 15 research labs, with access expanded in 2019 to all academic medical school constituents ... (Full article...)
|